Emanuele Rossi
MLxAnimal Communication @ Sapienza University

I’m a postdoctoral researcher in the Gladia group at Sapienza University of Rome, working with Emanuele Rodolà. My research explores how AI can help decode animal communication, focusing on how multimodal models can reveal what wild animals communicate to one another, what this tells us about their intelligence and consciousness, and how such understanding can reshape our relationship with the natural world.
Before turning my attention to animal communication, I worked at Vant AI, developing generative models for structural biology and drug discovery, and earned my PhD between Imperial College London and Twitter, supervised by Michael Bronstein and focusing on Graph Neural Networks. Earlier, I was part of Fabula AI, a deep learning startup for fake news detection that was acquired by Twitter. I hold degrees in Computer Science from Imperial College London and the University of Cambridge.
Outside research, I love scuba diving, hiking, and quiet moments in nature.
News
| Nov 10, 2025 | Our paper “Model Merging Improves Zero-Shot Generalization in Bioacoustic Foundation Models” has been accepted at the NeurIPS 2025 Workshop on AI for Animal Communication 🎉 |
|---|---|
| Nov 06, 2025 | I’m excited to share that I will join the Gladia Group at Sapienza University of Rome as a Postdoc, working with Emanuele Rodola. My research will explore how ML can help decode animal communication, and how this pursuit can, in turn, inspire the next generation of ML methods 🐬 |
| Mar 21, 2025 | Excited to finally share what we’ve been working on at Vant AI for the past year and a half: Neo-1, a unified model for all-atom structure prediction and generation of all biomolecules 🔬 |
| Jul 19, 2024 | The lack of large, high-quality datasets and robust evaluation is holding back ML in Drug Discovery. We are releasing Pinder (Protein-Protein) and Plinder (Protein-Ligand) to help bridge this gap and drive meaningful progress 🧬 |
| Jan 16, 2024 | I’m thrilled to announce that I’ve joined Vant AI as a Machine Learning Researcher. Vant combines a compelling ML vision with a proprietary data generation platform, focusing on the novel field of molecular glues. I’ll be developing generative models for structural biology to advance the drug discovery process 🚀 |
Selected Publications
- NeurIPS
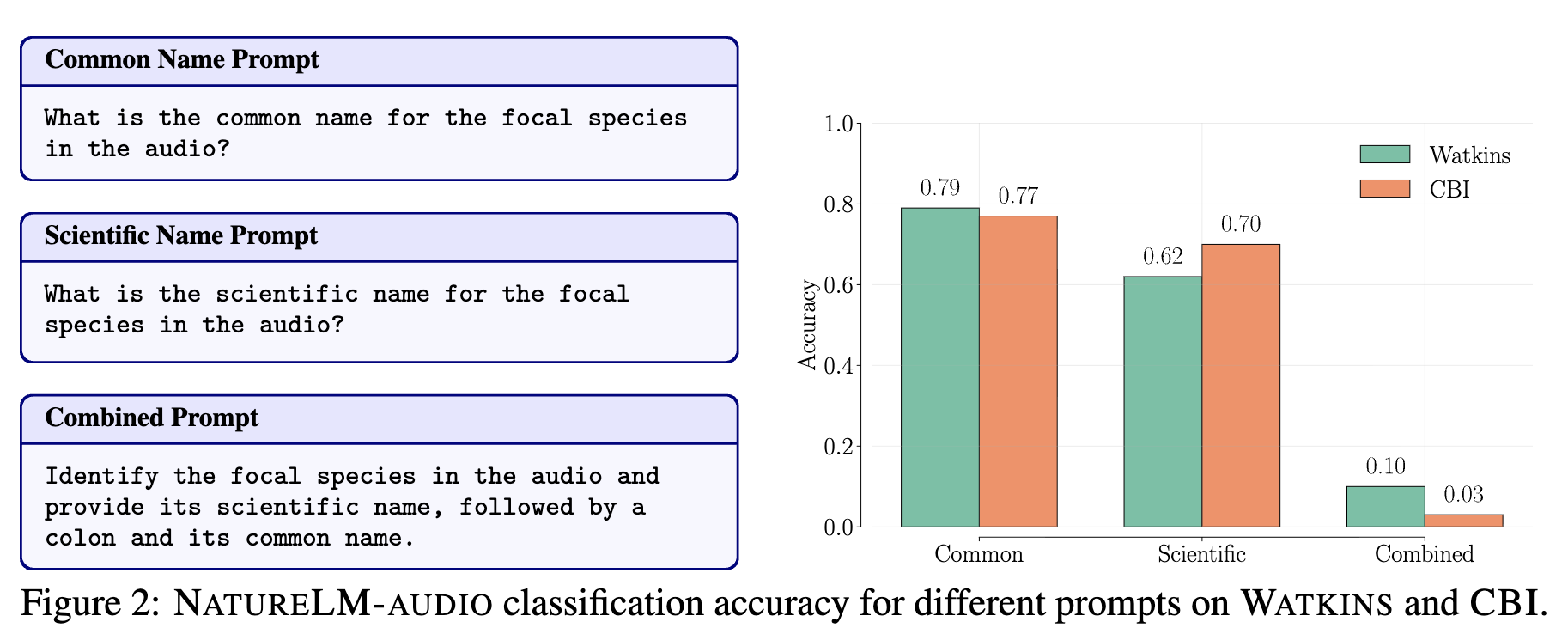 Model Merging Improves Zero-Shot Generalization in Bioacoustic Foundation ModelsNeurIPS Workshop on AI for Animal Communication, 2025
Model Merging Improves Zero-Shot Generalization in Bioacoustic Foundation ModelsNeurIPS Workshop on AI for Animal Communication, 2025 - LoG
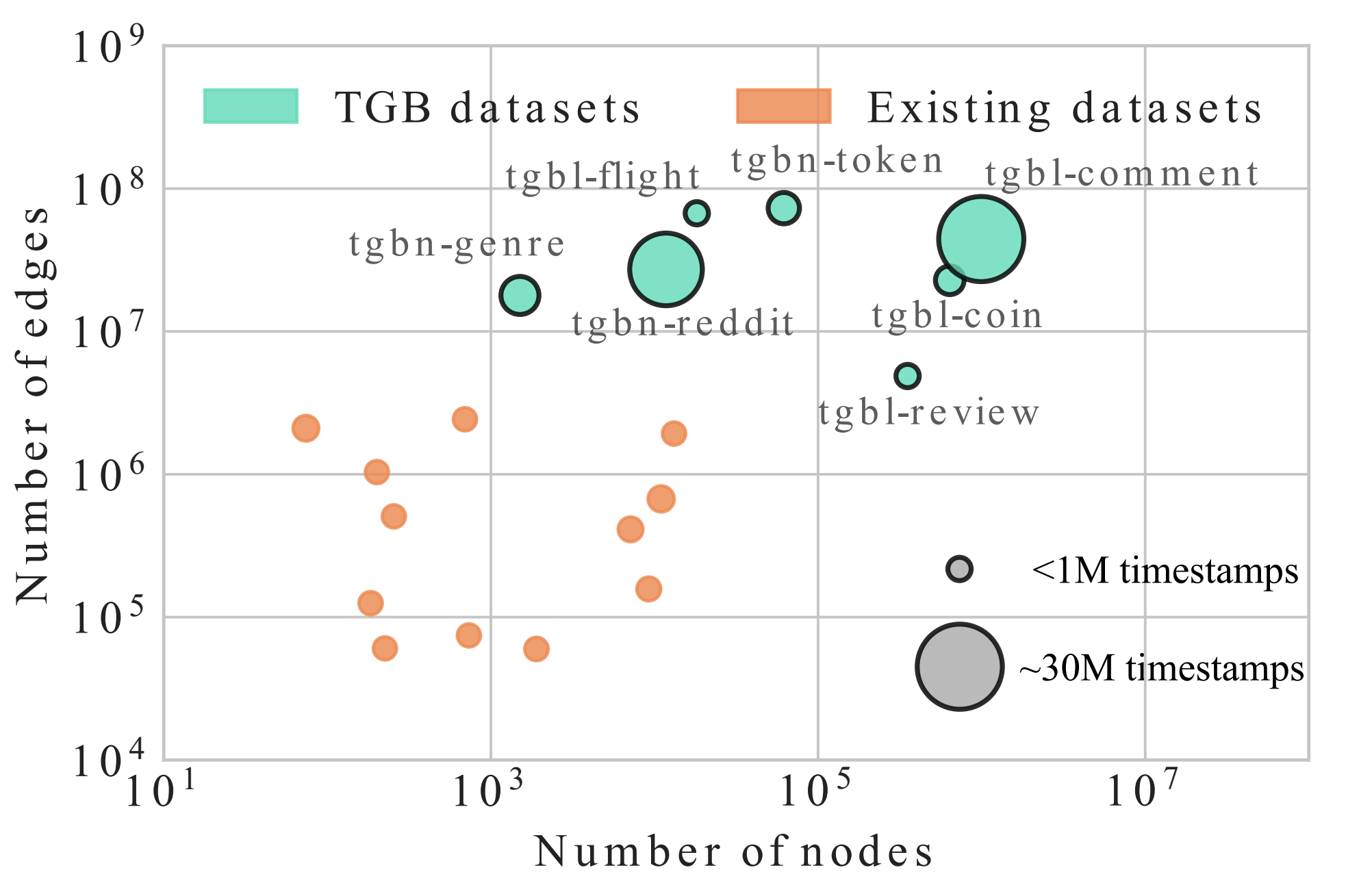
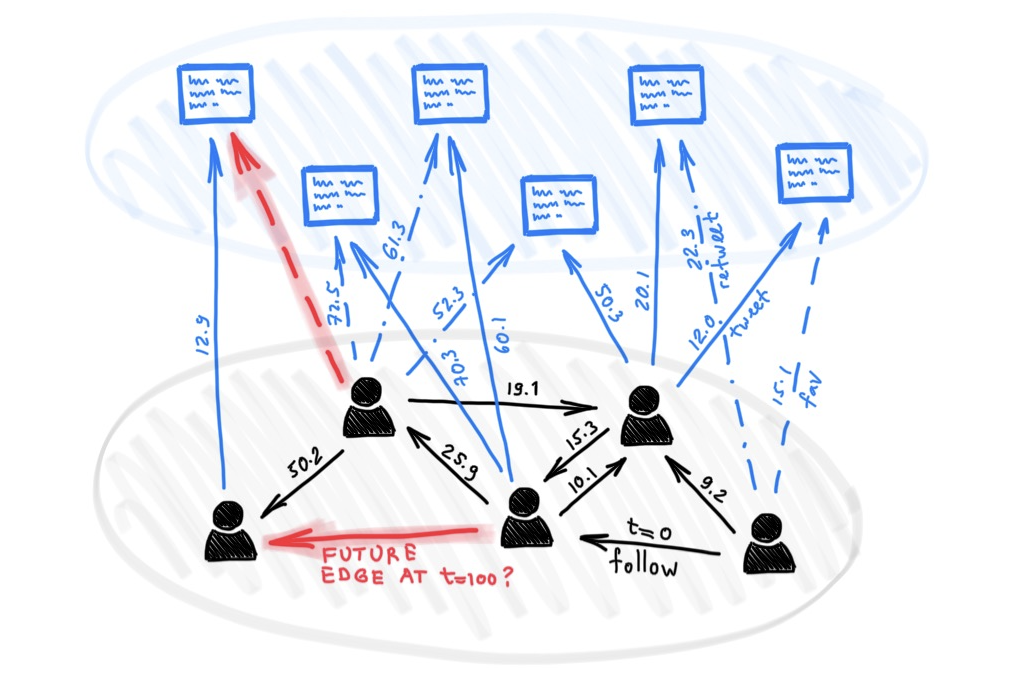
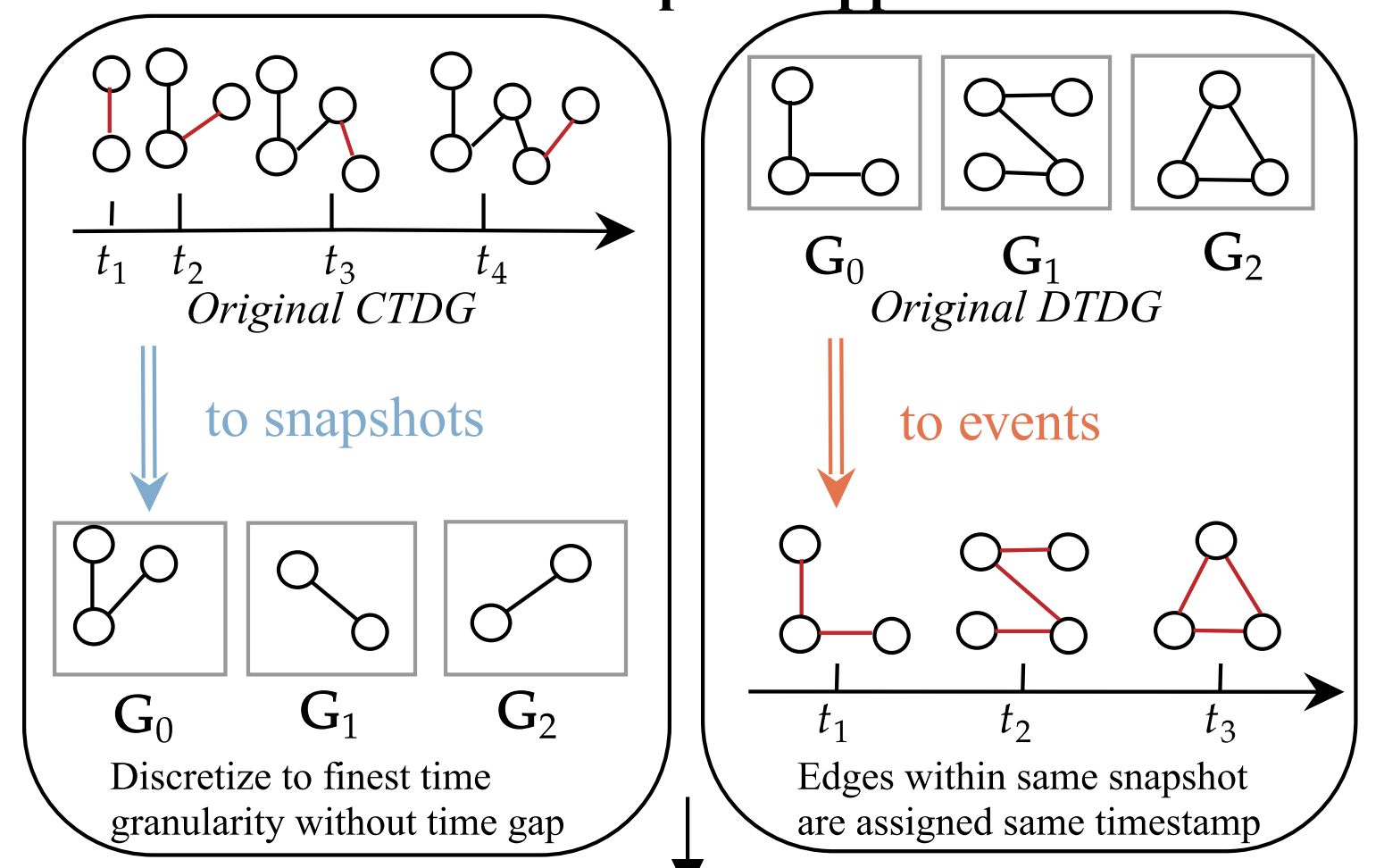 UTG: Towards a Unified View of Snapshot and Event Based Models for Temporal GraphsLearning on Graphs Conference (LoG), 2024
UTG: Towards a Unified View of Snapshot and Event Based Models for Temporal GraphsLearning on Graphs Conference (LoG), 2024